Orange installation guide
Introduction to data mining
Installation instructions for Orange Orange is developed by members of the Bioinformatics Lab, Ljubljana.Software installation instructions
Step 1: Python data mining libraries
For Mac OS X, Windows, or Linux.
Step 2: Testing the environment
Test the environment by running sample workflows. Instructions for all operating systems.
The following instructions will guide you through installation of various libraries and enviroments we will use during the tutorial.
Step 1: Mac OS X
In a nutshell
Download the latest version of Orange (i.e. a dmg installation file) from Orange website, install it on your system and continue with Step 2 of these instructions.
Detailed installation instructions
Download latest bundle for OSX from the Orange 3 download site .
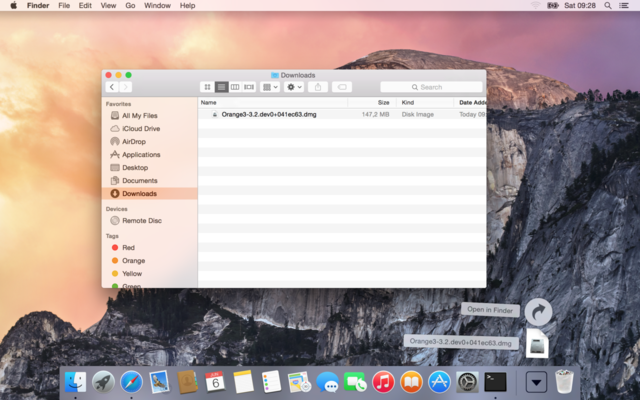
Your browser will most likely save the installation file (a file with a name similar to Orange3-3.2.dev0+e196459.dmg) to your Downloads folder.
Open the installation file by double clicking its icon.
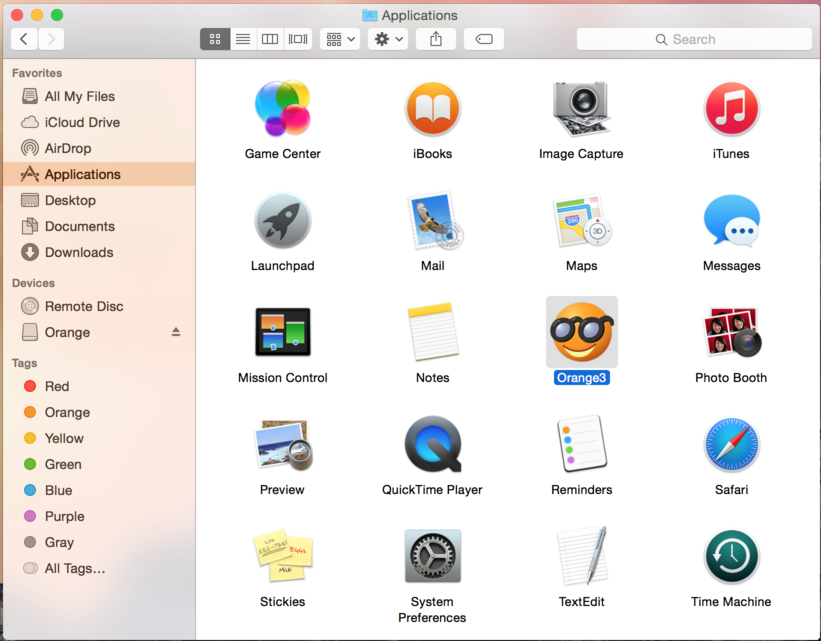
A window like the one shown below should open. Drag the Orange icon and release it over the Applications icon.Now you can open Orange from your Applications folder. Simply open the Applications folder, find the application named Orange, click its icon to select it, and open it by choosing Open from Finder's File menu. [You should use this procedure instead of a double click to open Orange, i.e. unregistered applications, for the first time.]
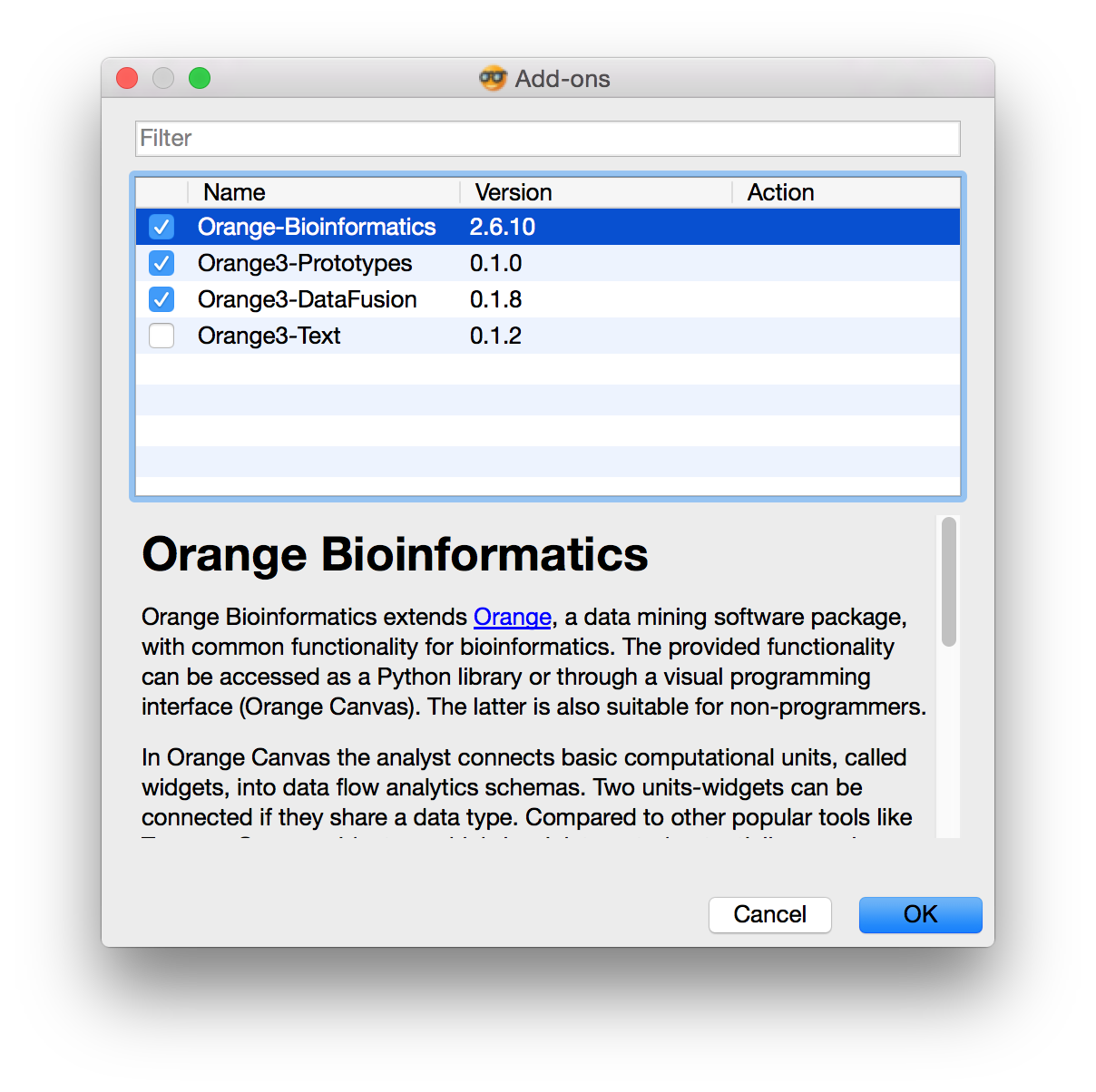
Install bioinformatics add-on by selecting Add-ons from menu Options. Dialog window like this one will appear.
Now check the Orange3-Bioinformatics add-on and press OK to install. If you don't have the Command Line Tools installed, the install procedure will prompt you to install them. Click Install button and click Agree with the Command Line Tools License Agreement. Upon installation completion, close and reopen Orange. Now you should see new Bioinformatics widgets in the left panel.
Continue with Step 2 of this installation tutorial.Step 1: Windows
In a nutshell
- Download and open Orange3 installation file.
- Follow installation wizard to install Orange 3 and its dependencies (Python 3.4, NumPy, etc.).
- Install Bioinformatics add-on.
- Restart Orange 3.
- Continue with Step 2.
Detailed installation instructions
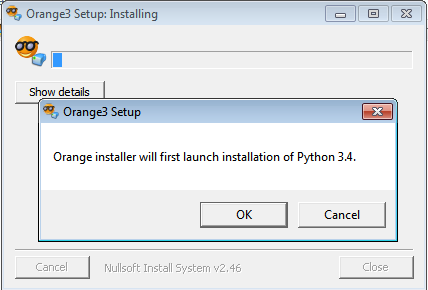
Download the installation file and save the file to an arbitrary location. Start the installation by double clicking the downloaded file. Choose Yes when asked whether you want to allow this program to make changes to your computer.
Setup will ask you to confirm installation of Python 3.4 on your computer. Press OK. If you already have Python 3.4 installed, this step is not needed.
Follow installation wizard to install Python 3.4. You can leave default settings and wait for installation to complete.
After installation of Python 3.4, the wizard will install Orange 3 and its dependencies. This step might take a while.
Double click on the Orange desktop icon to start the Orange application.
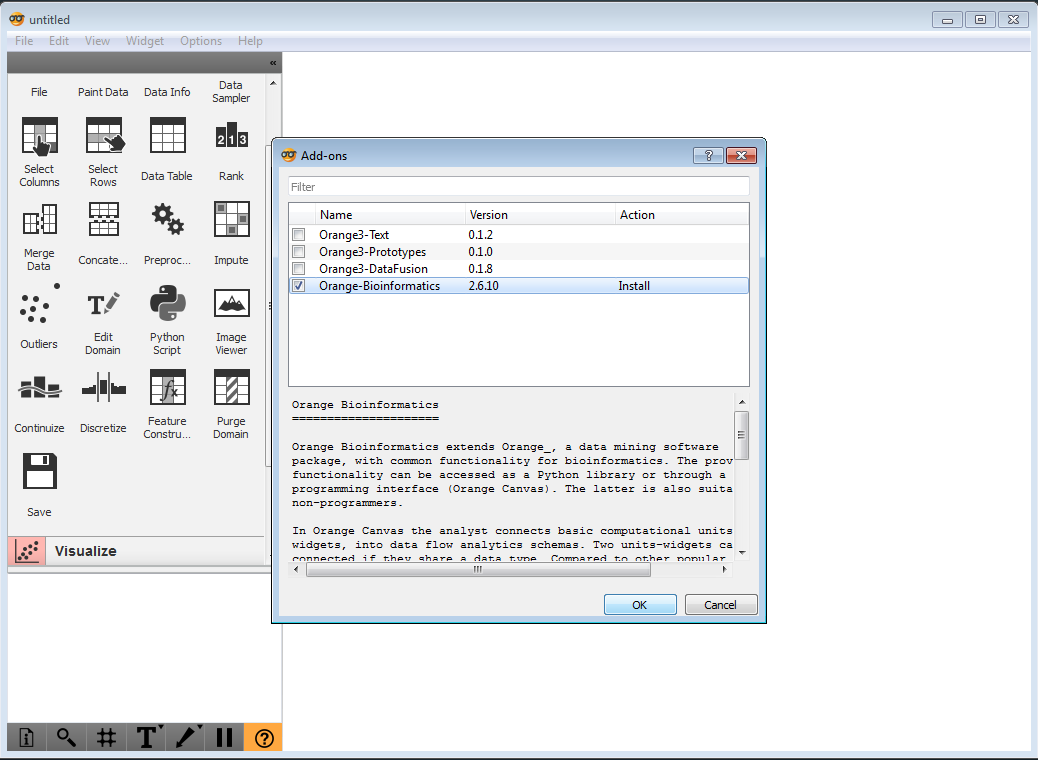
To install bioinformatics widgets, follow the menu Options->Add-ons. Check the box next to Orange3-Bioinformatics and press OK.
Alternatively, you can install Orange-Bioinformatics addon from command line using following command:
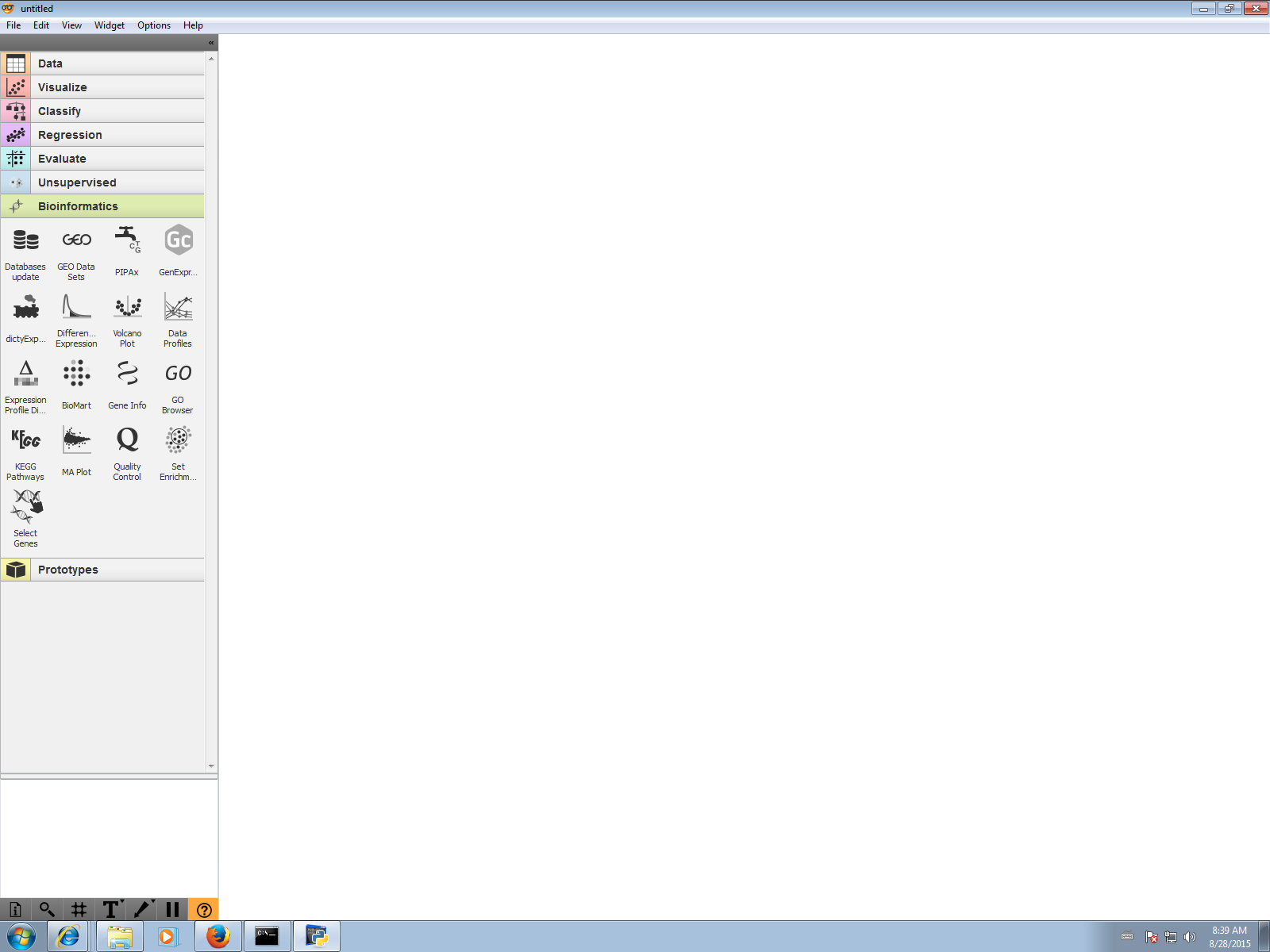
C:\Python34\Scripts\pip.exe install Orange-BioinformaticsFor changes to take effect you need to restart Orange. Close the main Orange window and then double click Orange Canvas icon on the desktop to restart Orange application. You should see a new Bioinformatics category with many widgets as shown below.
Install openpyxl using the Openpyxl install script .
Continue with Step 2 of this installation tutorial.Step 1: Linux
Ubuntu (tested on Ubuntu 14.04 and 14.10)
Automatic setup
Setup can be automated by using Ubuntu 14.04 setup script . Before the installation, move to directory, where you want Orange 3 to be installed. Following command will start installation, which usually takes about 15 minutes.
source setup-ubuntu.sh Install script will also create and enter new virtual environment for you, which you can reenter using following command:
source orange3env/bin/activate python -m Orange.canvasManual installation
Alternatively, you can choose manual installation. Open a terminal and run the following commands.
Install the necessary system packages:
sudo apt-get update
sudo apt-get install git python-pip python-virtualenv
python3-dev python3-numpy python3-scipy
python3-pyqt4 python-qt4-dev python3-sip-dev libqt4-dev
Note that Debian users might additionally need to install pkg-config package.
We will use Python3 and need to create a virtual environment for it:
mkdir orange3env
virtualenv -p python3 --system-site-packages orange3env
source orange3env/bin/activateInstall Orange and its requirements (this step will take about 15 minutes):
git clone https://github.com/biolab/orange3
cd orange3
pip install -r requirements.txt
pip install -r requirements-gui.txt
pip install -r requirements-sql.txt
python setup.py develop
cd ..
git clone https://github.com/biolab/orange-bio
cd orange-bio
python setup.py develop
Now you can open Orange with the following command
python -m Orange.canvasopenSUSE (tested on openSUSE 13.2)
Automatic setup
Install can be automated by using openSUSE setup script . Before the installation, move to directory, where you want Orange 3 to be installed. Following command will start installation, which usually takes about 15 minutes.
source setup-suse.sh Install script will also create and enter new virtual environment for you, which you can reenter using following command:
source orange3env/bin/activate python -m Orange.canvasManual installation
Alternatively, you can choose manual installation. Open a terminal and run the following commands.
Install the necessary system packages:
sudo zypper refresh
sudo zypper install git python-virtualenv python3-devel gcc gcc-c++
python3-numpy python3-numpy-devel python3-scipy
python3-qt4-devel
We will use Python3 and need to create a virtual environment for it:
mkdir orange3env
virtualenv -p python3 --system-site-packages orange3env
source orange3env/bin/activateInstall Orange and its requirements (this step will take about 15 minutes):
git clone https://github.com/biolab/orange3
cd orange3
pip install -r requirements.txt
pip install -r requirements-gui.txt
pip install -r requirements-sql.txt
python setup.py develop
cd ..
git clone https://github.com/biolab/orange-bio
cd orange-bio
python setup.py developNow you can open Orange with following command
python -m Orange.canvasStep 2: Testing the environment
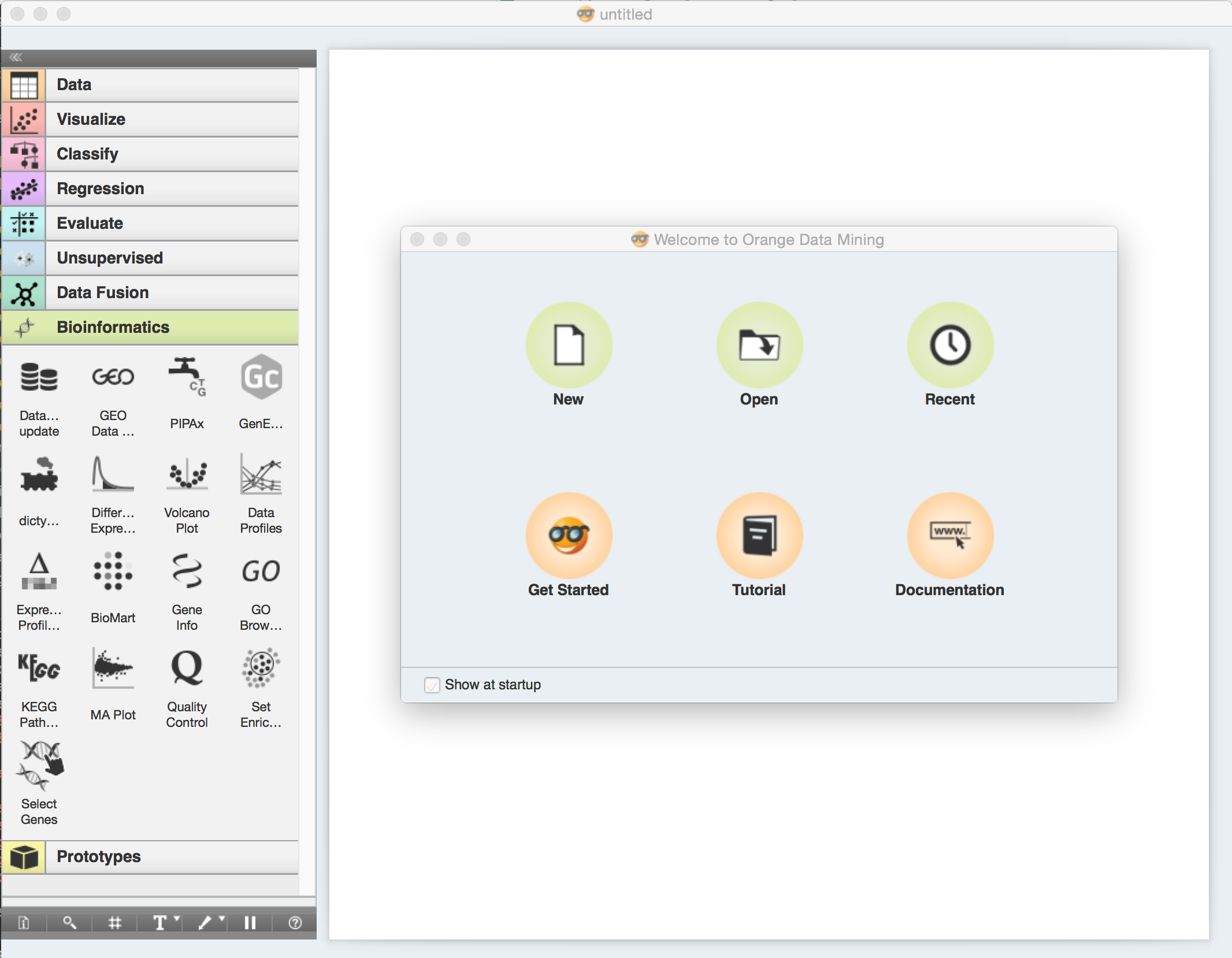
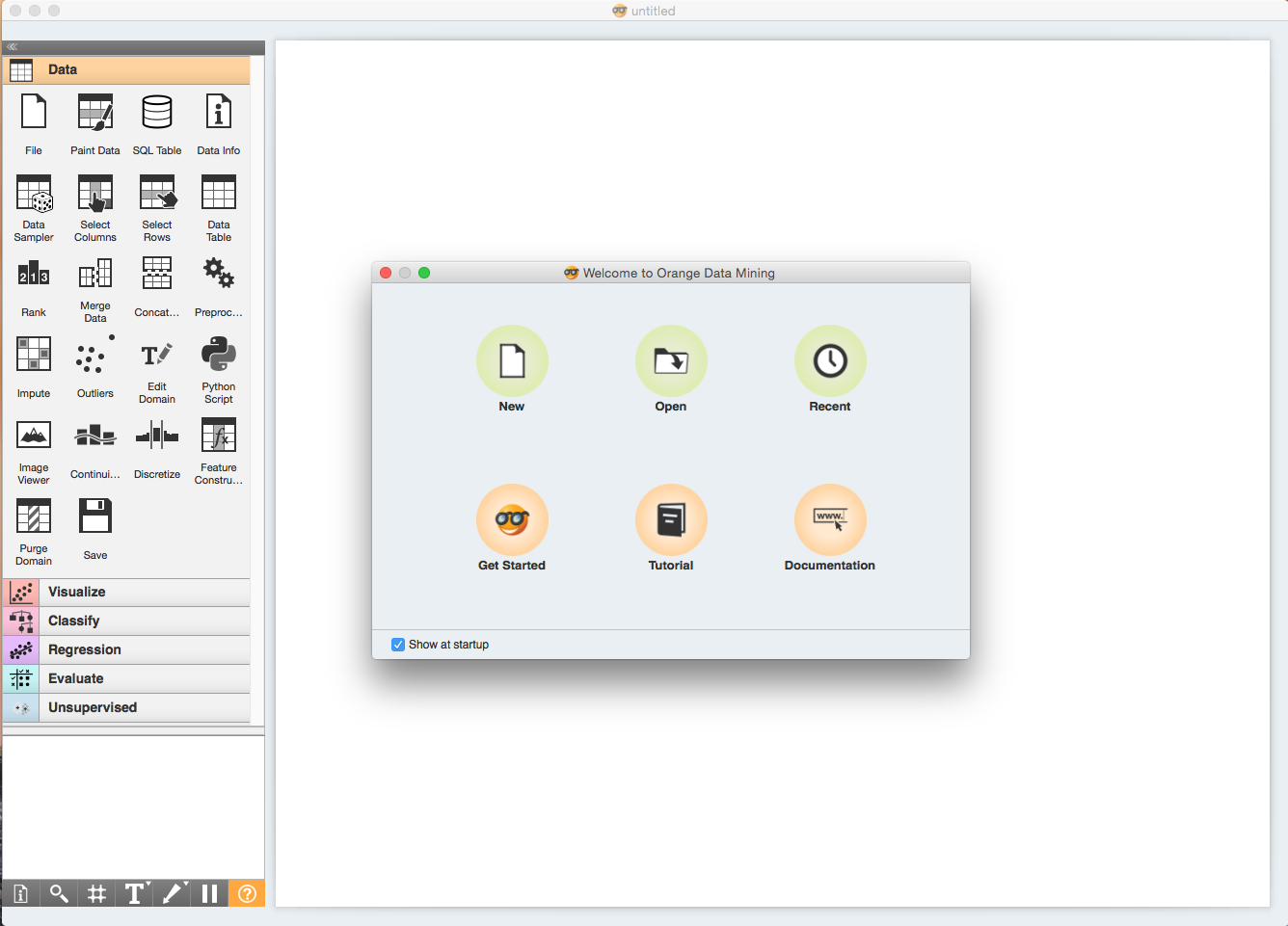
You should see a welcome screen as the one shown below when you start Orange. Choose Tutorial. Alternatively, you can reach tutorials by selecting Tutorials from the Help menu.
A window like the one shown below will open.
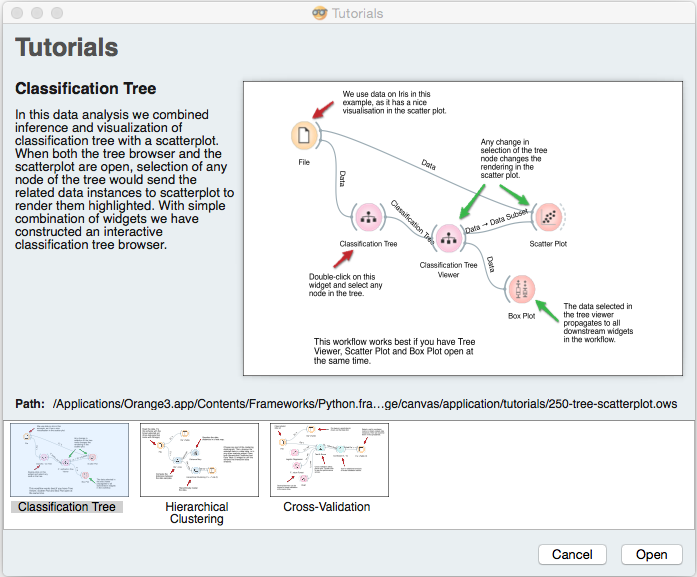
Select the Classification tree workflow and click Open to open it.
That's it! We are looking forward to having you attend the course.